Module 6
1. Module 6
1.41. Page 2
Module 6—Mendelian Genetics: The Transmission of Traits to the Next Generation
 Explore
Explore
 Read
Read
Recall from Lesson 6 on sex–linked inheritance that Thomas Hunt Morgan provided experimental evidence that genes occur on chromosomes. This discovery immediately created a new idea from Mendel’s laws: the idea that traits could tend to move together if they were on the same chromosome.
Mendel did not find that any of the seven traits he studied moved together. Each trait had no relation to another: each trait assorted independently. The traits that Mendel studied in peas were found on different chromosomes, so we would expect them to move independently.

However, many traits in organisms are coded on the same chromosome. These genes tend to move together, and are thus called linked genes. When first considering linked genes, you may think that these genes always move together; however, that is not the case. Even though they are on the same chromosome, there is an opportunity to exchange pieces of homologous chromosomes during meiosis. This process is called crossing over.
The farther apart two genes are on a chromosome, the greater the number of crossover events that will occur between them, and it is less likely that the two traits will move together. The number of crossover events directly relates to distance on a chromosome. Read “Crossing Over and Inheritance” to the end of “Chromosome Mapping” on pages 599 to 601 of your textbook.
 Watch and Listen
Watch and Listen
Watch the following segments of the video “Chromosomal Basis of Inheritance: The Choreography of Generations.” Remember that you can ask your teacher for a username and password to access the video.
- “Bio Reports: Inheritance”
- “Dyhybrid Crosses”
- “Bio Bit: Genetic Diversity”
- “Bio Discovery: Gene Linkage”
- “Gene Linkage”
- “Bio Discovery: Crossing Over”
- “Bio Probe: Gene Mapping”
- “Bio Bit: Chromosome and Gene Mapping”
- “Bio Bit: Barbara McClintock”
Remember to add the new terms to your flash cards.
 Try This
Try This
Apply your understanding to the following questions and save your work in your course folder.
parental type: in a linkage cross, if the offspring look like either parent, then they are parental types (e.g., if we assume A and B are linked together and a and b are linked together in the linkage cross AaBb X aabb, any offspring that are AaBb or aabb are parental types)
non-parental type: in a linkage cross, any offspring of the above cross that don’t look like the parents (Aabb or aaBb) are non-parental types, are the results of crossing over, and are recombinants
TR 1. How would the phenotypic ratio of the cross GgRr × ggrr be different for independent assortment than for linked genes?
TR 2. What is meant by parental types of chromosomes?
TR 3. What is meant by non-parental types of chromosomes?
TR 4. How does recombination frequency relate to map distance?
recombination frequency: the number of recombinant offspring divided by the total number of offspring X 100; expressed as a percentage; used as a map distance between the two genes (e.g., the recombination frequency was 32%, therefore the two genes are 32 map units apart on the chromosome)
map distance: how far apart (in centimorgans) two genes are on a chromosome; the recombination frequency from a linkage cross is used as the map distance between two genes (e.g., 44% of the offspring of the linkage cross were recombinants, therefore the two genes are 44 map units or 44 cM apart)
 Module 6: Lesson 9—Lab: Mapping Chromosomes
Module 6: Lesson 9—Lab: Mapping Chromosomes
In this lab you will practise creating your own map by completing Part A of “Thought Lab 17.1: Mapping Chromosomes” on page 602 of your textbook. This work will become part of your assignment for assessment in this lesson.
Retrieve your copy of Module 6: Lesson 9 Assignment that you saved to your computer earlier in this lesson. Complete Part A of the “Thought Lab”. Save your work in your course folder. You will receive instructions later in this lesson on when to complete your assignment and submit it to your teacher.
Then complete Part B of “Thought Lab 17.1: Mapping Chromosomes” on page 602. Discuss your work in Part B with your teacher and save your responses to your course folder.
 Self-Check
Self-Check
Determining recombinant types in crosses and calculating recombination frequency is challenging. However, maps can be easily created from those frequencies. To help you practice the principles of gene mapping, answer the following questions.
SC 1. If there were 50 recombination phenotypes in 250 offspring, what is the map distance between the linked alleles?
recombination: during crossing over in prophase I of meiosis, genes from non-sister chromatids trade places, making new allele combinations in the gametes (e.g., if A is linked to B and a to b, then any gametes that are Ab or aB are the results of recombination or crossingover)
map unit: the unit used is the centimorgan (cM); because the recombination frequency is a percentage, the largest map distance is 100 cM
crossover percentage: same as recombination frequency
SC 2. A three-point test cross is performed to identify the locus of each of three alleles in relation to one another. The results were as follows:
- AB recombinations = 225
- BC recombinations = 165
- AC recombinations = 60
- Parental linkages = 550
- Total offspring = 1000
Show the positions and map the distances apart for each allele (A, B, and C) on a chromosome. Calculate the map units, then draw the chromosome. You will encounter the term crossover percentage in the answer provided.
 Self-Check Answers
Self-Check Answers
SC 1. Crossover % = 50/250 × 100 = 20%
Since 1% crossover represents 1 map unit, 20% crossover would mean 20 map units apart.
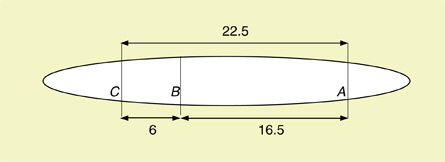
SC 2. The crossover percentages and map units are as follows:
- AB = 225/1000 × 100% = 22.5% or 22.5 map units
- BC = 165/1000 × 100% = 16.5% or 16.5 map units
- AC = 60/1000 × 100% = 6.0% or 6.0 map units
Therefore, the mapped chromosome would look like this: